Translational Genomics & Bioinformatics Lab
Who We Are
The Laboratory for Translational Genomics and Bioinformatics is in the Department of Pathology and Laboratory Medicine in the David Geffen School of Medicine at UCLA. We are also affiliated with the Quantitative and Computational Bioscience (QCBio) Institute, and the Bioinformatics Interdepartmental Ph.D. Program (IDP).
What We Do
Precision Medicine
Precision medicine is the use of a patient's genomic information to offer more precise and targeted treatment. NGS technology is revolutionizing disease diagnosis, monitoring, and treatment. There has never been a better time for computational biologists to make real contributions to the practice of health care.
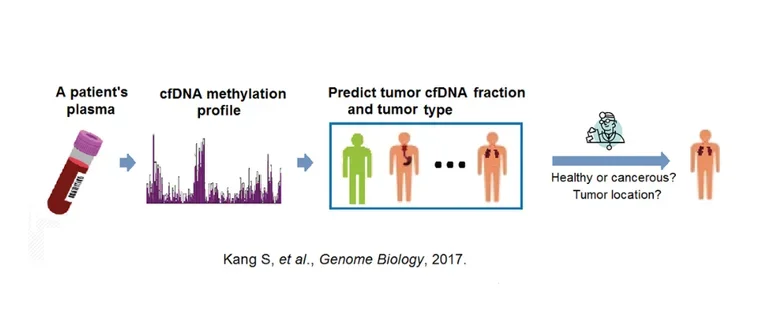
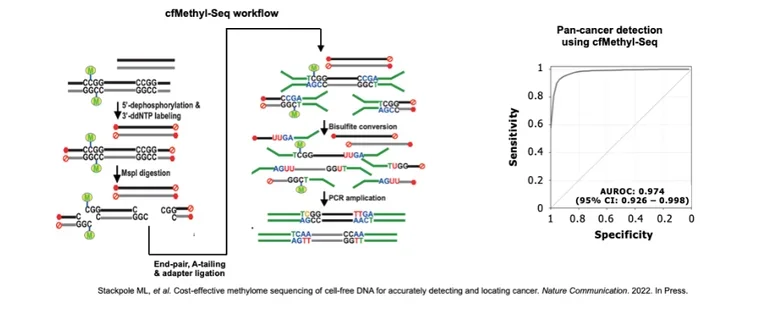
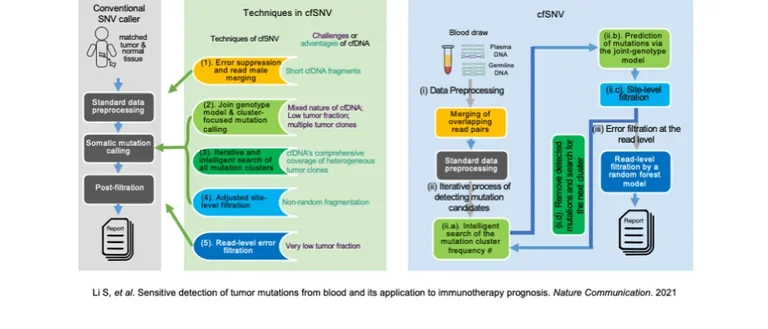
For example, over the past year we have been working on non-invasive methods of cancer diagnosis and monitoring based on “liquid biopsy”: the detection of circulating free DNA (cfDNA) released by tumors into the blood.
Big Biodata Analysis
Big biodata analysis has been a focus of our lab for more than a decade. The rapid accumulation of genomics data in public repositories has created an urgent need for methods that can effectively integrate large datasets generated by different platforms and modalities. In fact, the “big data” challenge of biology is more a “complex data” challenge, since even petabytes of many NGS datasets can be significantly compressed in size through preprocessing, but not in its biological complexity and platform heterogeneity. We have developed a series of methods to address this challenge. Our efforts include both horizontal integration (combining datasets of the same type, such as hundreds or even thousands of gene expression datasets) and vertical integration (combining datasets of different types, such as Hi-C, epigenomic, expression, and textual data).
Some highlights of our past accomplishments are: we made the first attempt to transform public expression repositories into a diagnosis database (using a Bayesian framework); we were the first to recognize the problem of pattern mining across large many massive networks, and developed a suite of algorithms address this challenge; we developed a series of method for the analysis of multi-modality genomics profilings of same samples to reveal the coupling of multi-level gene regulations.